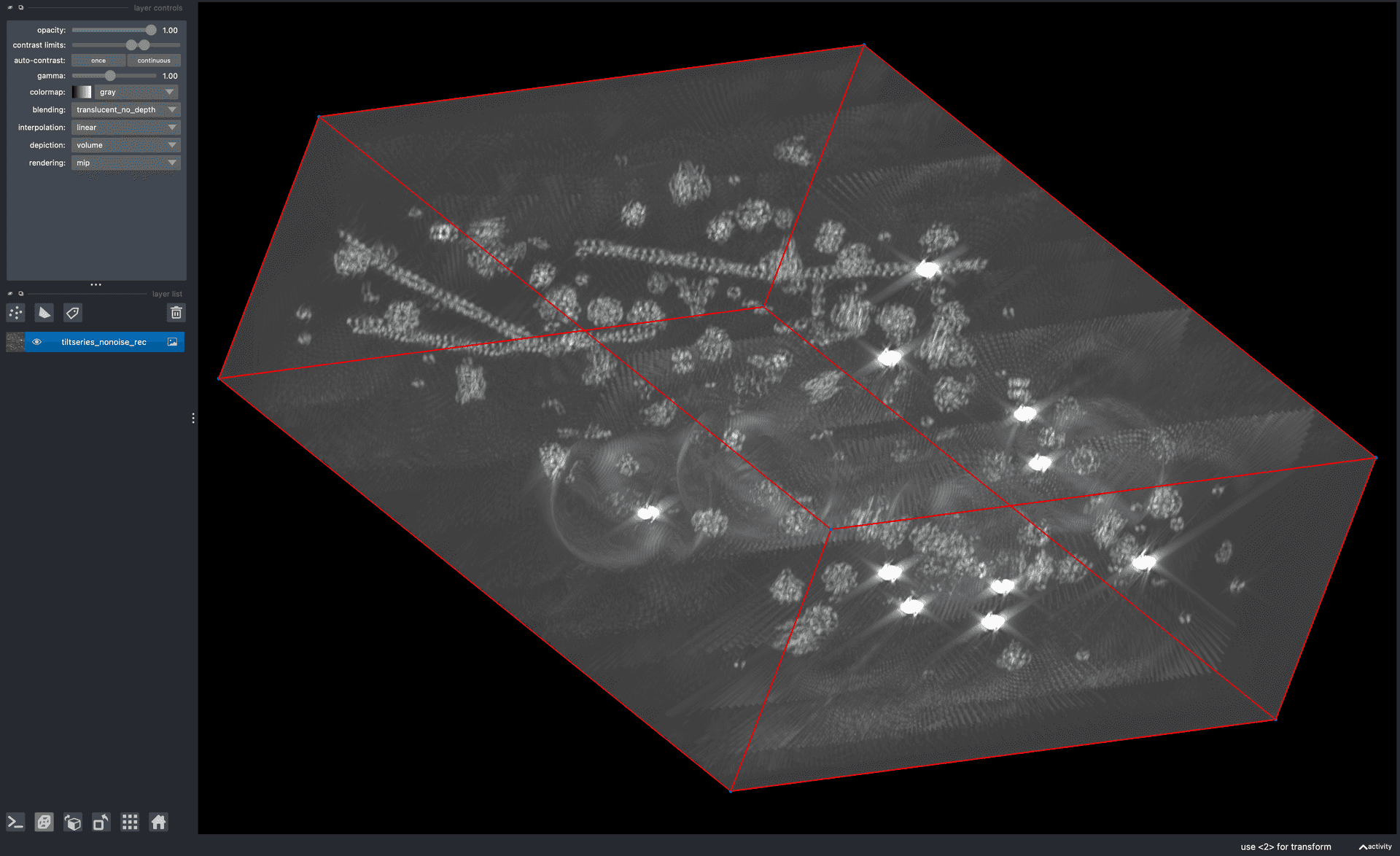
tem simulator script.
Tags
templatejavaimagej2
Citation
H. Rullgård, L.-G. Öfverstedt, S. Masich, B. Daneholt, and O. Öktem, Simulation of transmission electron microscope images of biological specimens, Journal of Microscopy, 242 (2011). Provided by https://github.com/MPI-Dortmund/tem-simulator-scripts which is part of TomoTwinhttps://tem-simulator.sourceforge.net/
Solution written by
Kyle Harrington
License of solution
MIT
Arguments
--pdbs
Path to PDB files (default value: /Users/kharrington/git/MPI-Dortmund/tem-simulator-scripts/pdbs/*.pdb)
--output
Output directory (default value: out_sim_tomo_1)
--random_seed
Random seed (default value: 10)
--pdbs_fil
Path to filament PDB files (default value: /Users/kharrington/git/MPI-Dortmund/tem-simulator-scripts/resources/filament_files/*.pdb)
--settings_fil
Path to settings file (default value: /Users/kharrington/git/MPI-Dortmund/tem-simulator-scripts/resources/filament_files/*.json)
--nsubs
Number of substitutions (default value: 100)
--dose
Dose (default value: 15000)
--fiducialsize
Size of fiducials (default value: PARAMETER_VALUE)
--vesiclesize
Size of vesicles (default value: PARAMETER_VALUE)
--nvesicle
Number of vesicles (default value: PARAMETER_VALUE)
--nfiducial
Number of fiducials (default value: PARAMETER_VALUE)
--defocus_lower
Lower limit of defocus (default value: PARAMETER_VALUE)
--defocus_upper
Upper limit of defocus (default value: PARAMETER_VALUE)
--thickness
Thickness (default value: PARAMETER_VALUE)
--s1
Early abort flag (default value: 0)