- ImageJ2 templateAn Album solution template generating an ImgLib2 image with random values in ImageJ2.Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T., & Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics, 18(1).
![Dummy cover image.]()
- Parent environment for some napari cryoet tools.A parent environment for cryoet solutionsKyle Harrington.
- Parent environment for membrain_seg.A parent environment with git main of membrain_segteamtomo.
- n5-utils from Saalfeld LabAn Album solution for using Saalfeld Lab's n5 utils.Saalfeld lab.
- Java-based software parent solutionParent of Java-based software solutions: Fiji, ImageJ, ImgLib2, etc.SciJava community.
![Cover image for java-based software.]()
- Parent environment for pytme.A parent environment with git main of pytmeKosinskiLab.
- preprocessing for pytme.A command-line preprocessing tool for pytmeKosinskiLab.
- preprocessing gui for pytme.A napari-based preprocessing tool for pytmeKosinskiLab.
![Cover image for pyTME's preprocess gui. Shows an instance of napari running the plugin. Image comes from https://kosinskilab.github.io/pyTME/quickstart/preprocessing.html.]()
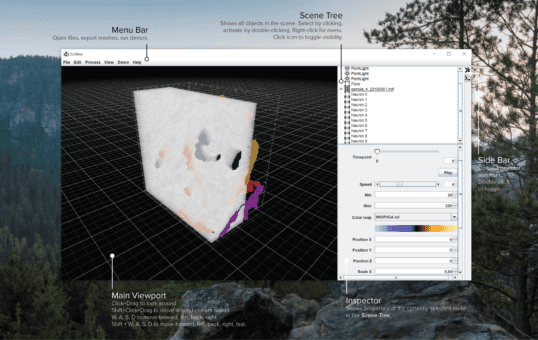
- sciviewsciview is a 3D/VR/AR visualization tool for large data from the Fiji communitysciview team.
![Cover image for sciview. Shows a schematic of the sciview UI with an electron microscopy image that contains segmented neurons.]()
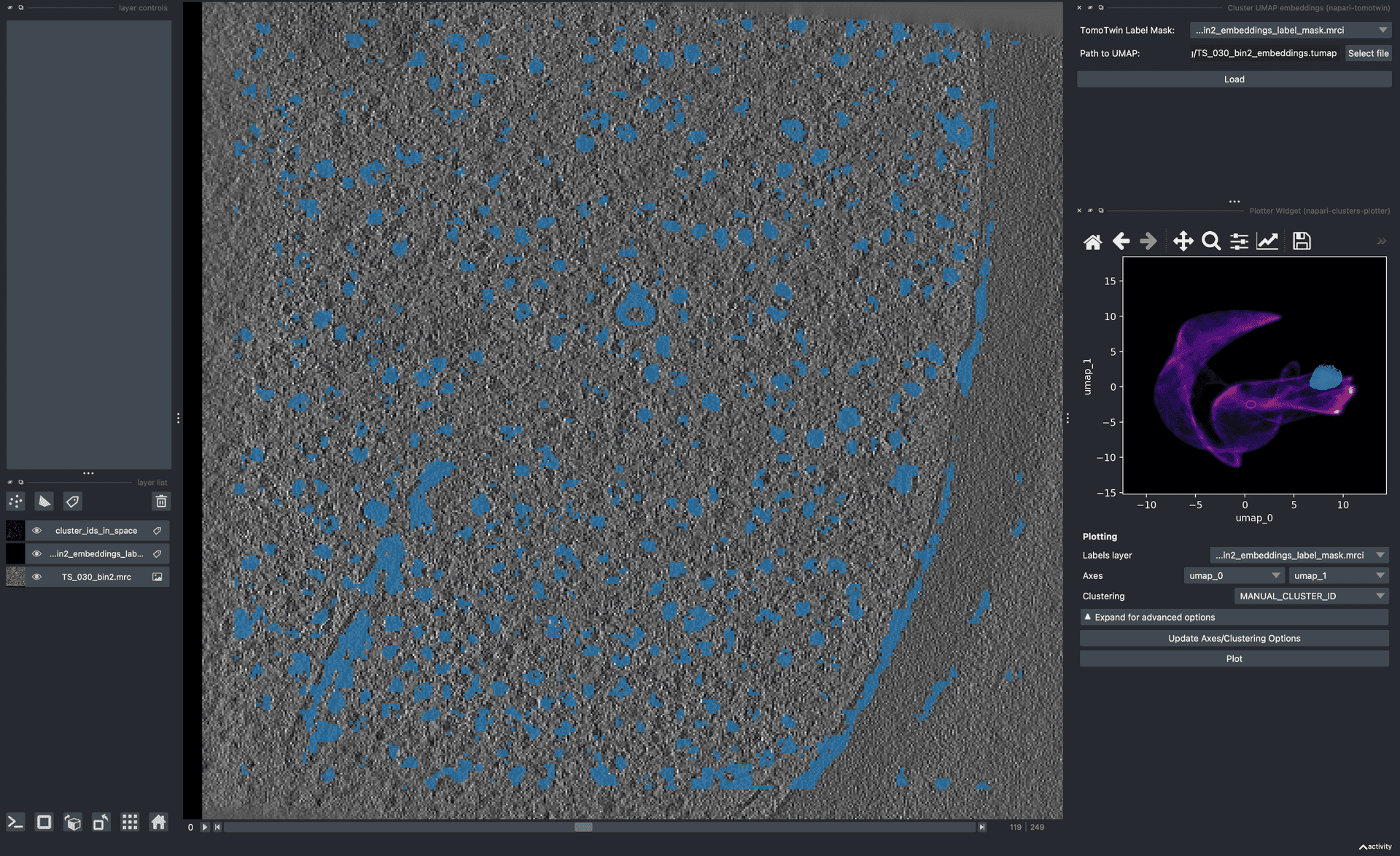
- TomoTwin demo on cz cryoet data portalTomoTwin on an example from the czii cryoet dataportal.Rice, G., Wagner, T., Stabrin, M. et al. TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01878-z.
![Cover image for TomoTwin tutorial 2 applied to data from cryoet-data-portal TS_030 from doi:10.1038/s41592-022-01746-2. The image shows a highlighted region of embedding space that covers some particles in the tomogram.]()
- TomoTwin tutorial 2.A tutorial for TomoTwinRice, G., Wagner, T., Stabrin, M. et al. TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01878-z.
![Cover image for TomoTwin. The image comes from the TomoTwin documentation: https://tomotwin-cryoet.readthedocs.io.]()
- AreTomo2: Automated Tomographic Alignment and ReconstructionAreTomo2 is a multi-GPU accelerated software package that automates motion-corrected marker-free tomographic alignment and reconstruction. It includes robust GPU-accelerated CTF estimation, offering fast, accurate, and easy integration into subtomogram processing workflows. AreTomo2 is capable of on-the-fly reconstruction of tomograms and CTF estimation in parallel with tilt series collection, enabling real-time sample quality assessment and collection parameter adjustments.Zheng, S., Wolff, G., Greenan, G., Chen, Z., Faas, F.G., Bárcena, M., Koster, A.J., Cheng, Y. and Agard, D.A., 2022. AreTomo: An integrated software package for automated marker-free, motion-corrected cryo-electron tomographic alignment and reconstruction. Journal of Structural Biology: X, 6, p.100068.
![Example of an AreTomo2 reconstructed tomogram, showcasing the capabilities of this automated alignment and reconstruction software.]()
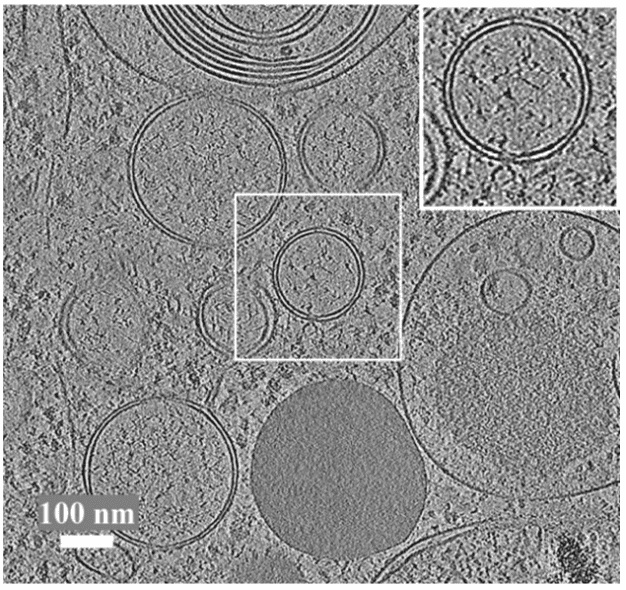
- cryOLO demo on cz cryoet data portalcryolo on an example from the czii cryoet dataportal.Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D. and Quentin, D., 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology, 2(1), p.218..
![Cover image for crYOLO from https://cryolo.readthedocs.io/en/stable/index.html.]()
- CTFFIND4: Fast and Accurate Defocus EstimationCTFFIND4 is a software package for accurate and fast estimation of defocus and astigmatism in electron micrographs. It is widely used in cryo-electron microscopy for preprocessing images before further analysis.Rohou, A., & Grigorieff, N. (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology, 192(2), 216-221.
![Image from https://grigoriefflab.umassmed.edu/ctffind4.]()
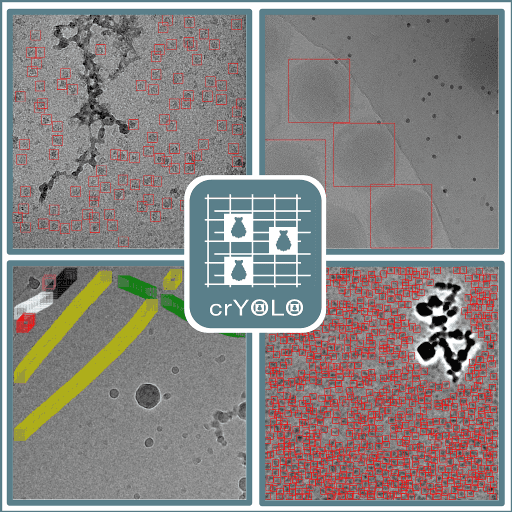
- crYOLO traintraining command for crYOLO.Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D. and Quentin, D., 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology, 2(1), p.218..
![Cover image for crYOLO from https://cryolo.readthedocs.io/en/stable/index.html.]()
- crYOLO predictprediction command for crYOLO.Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D. and Quentin, D., 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology, 2(1), p.218..
![Cover image for crYOLO from https://cryolo.readthedocs.io/en/stable/index.html.]()
- crYOLO evaluationevaluation command for crYOLO.Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D. and Quentin, D., 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology, 2(1), p.218..
![Cover image for crYOLO from https://cryolo.readthedocs.io/en/stable/index.html.]()
- crYOLO configconfig command for crYOLO.Wagner, T., Merino, F., Stabrin, M., Moriya, T., Antoni, C., Apelbaum, A., Hagel, P., Sitsel, O., Raisch, T., Prumbaum, D. and Quentin, D., 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications biology, 2(1), p.218..
![Cover image for crYOLO from https://cryolo.readthedocs.io/en/stable/index.html.]()
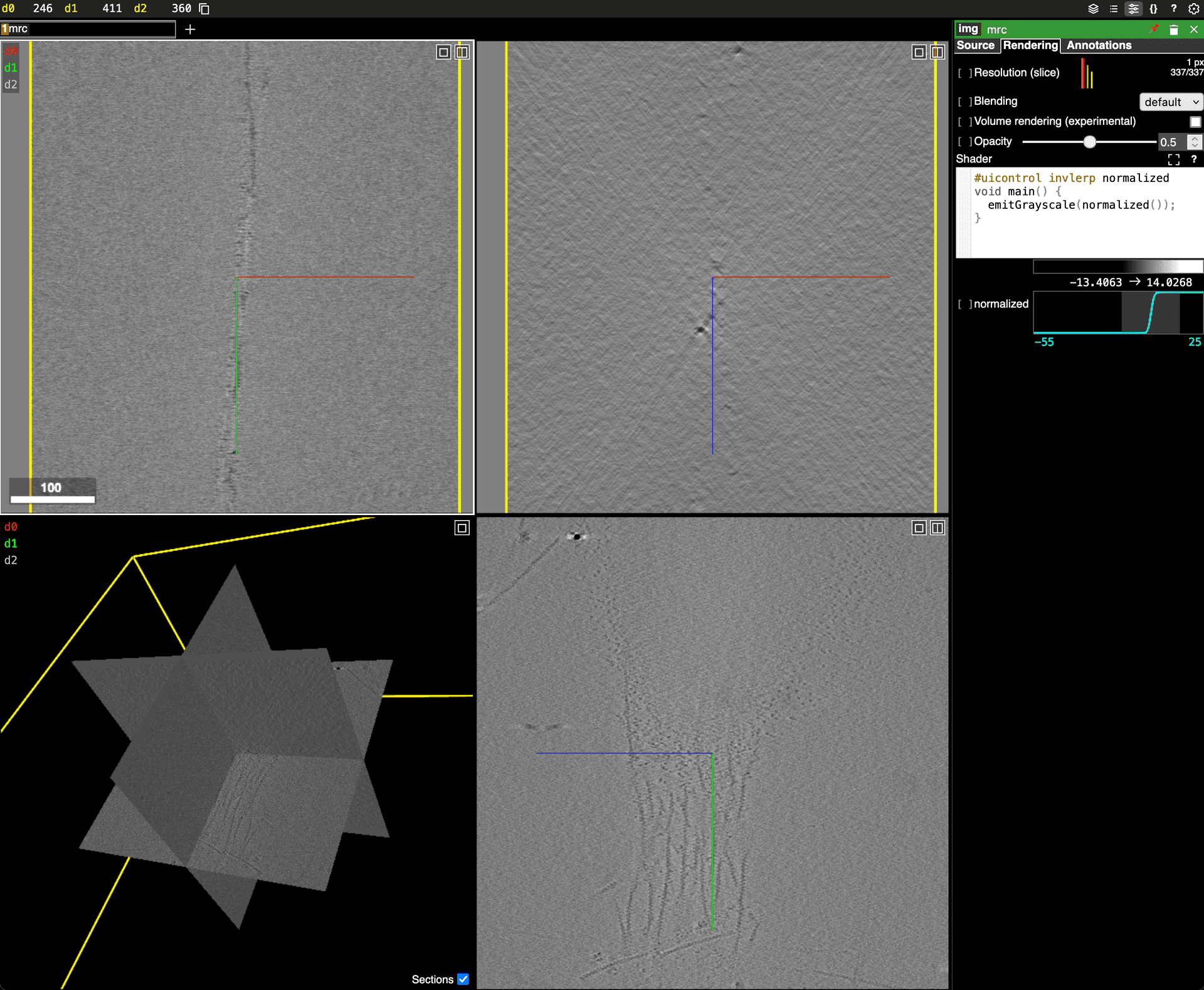
- View a MRC file with neuroglancerNeuroglancer viewer for MRC files.Neuroglancer by Google folks.
![Example of Neuroglancer visualizing EMPIAR-10548 dataset.]()
- View a remote MRC file with neuroglancerNeuroglancer viewer for MRC files that runs on a remote system.Neuroglancer by Google folks.
![Example of Neuroglancer visualizing EMPIAR-10548 dataset.]()
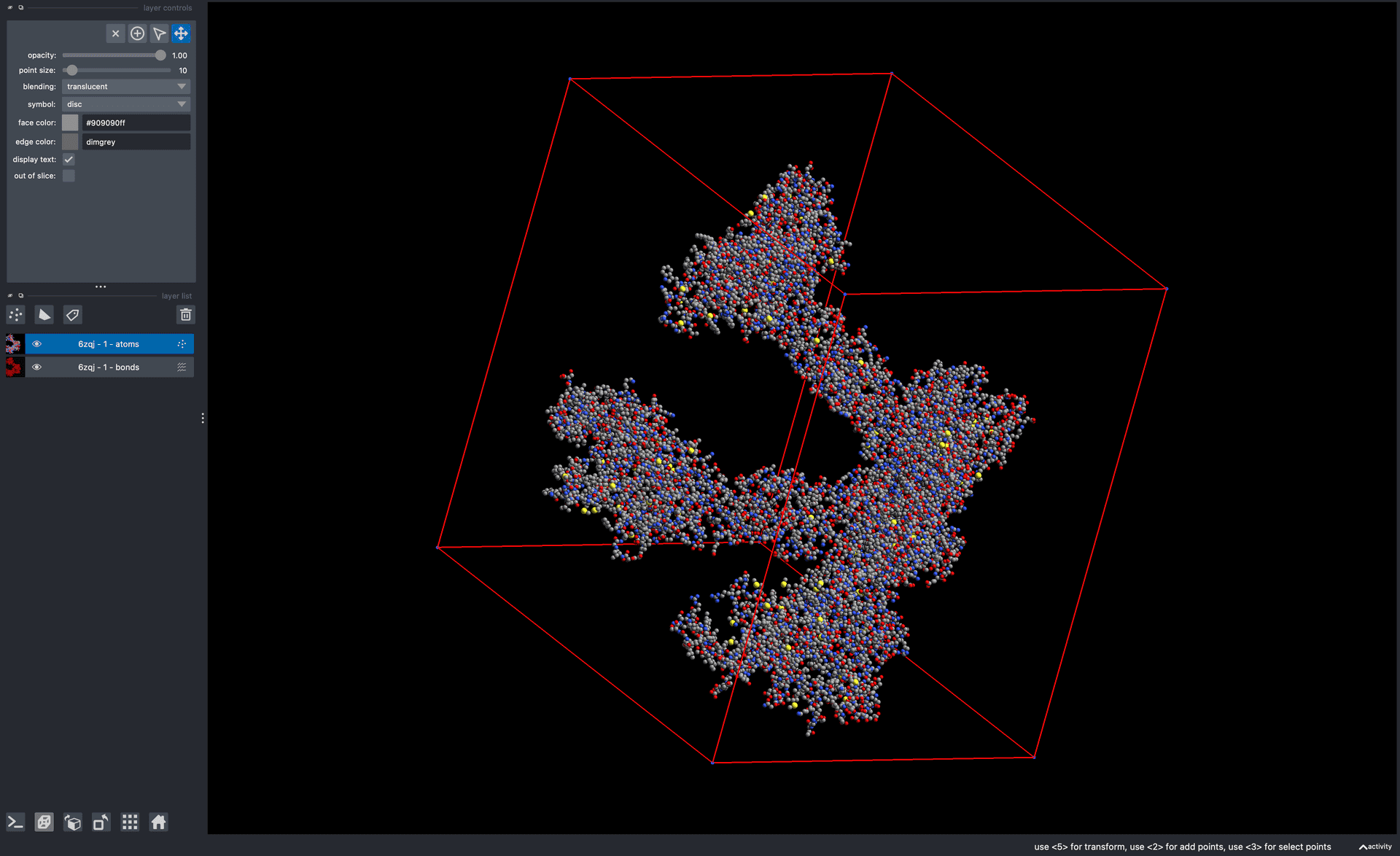
- PDB File DownloaderA utility to download PDB files from a list of PDB IDs.
![Visualization of PDB:6zqj with Lorenzo Gaifas' napari-molecule-reader plugin.]()
- tem-simulator-scripttem simulator script.H. Rullgård, L.-G. Öfverstedt, S. Masich, B. Daneholt, and O. Öktem, Simulation of transmission electron microscope images of biological specimens, Journal of Microscopy, 242 (2011). Provided by https://github.com/MPI-Dortmund/tem-simulator-scripts which is part of TomoTwin
![Screenshot of the volume render of a reconstruction created with this solution. Visualized in napari.]()
- album tutorial for czii in 2023This solution runs a Jupyter-based album tutorial presentation.
- Generate an embedding with TomoTwin for a mrcTomoTwin on an example from the czii cryoet dataportal.Rice, G., Wagner, T., Stabrin, M. et al. TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01878-z.
- CryoCanvas backendTBD.TBD.
- Tomogram fingerprintCompute the dataset fingerprints of a cryoET tomogram.Kyle. Also check out team tomo
- Parent environment for a big python environment for cryoET.A parent environment for a big python environment for cryoETKyle. Also check out TeamTomo
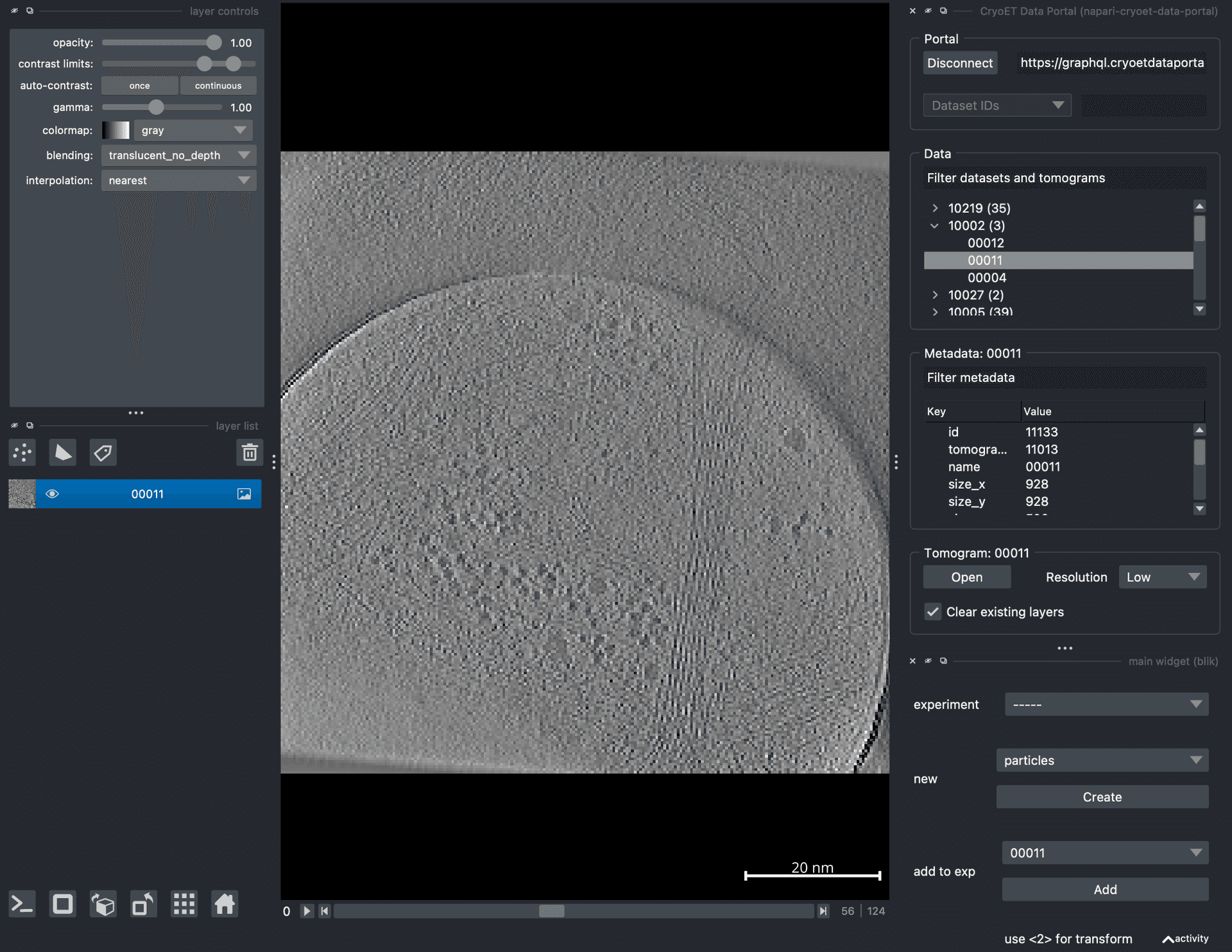
- Parent environment for a big napari environment for cryoET.A parent environment for a big napari environment for cryoETKyle. Also check out TeamTomo
![Screenshot of napari showing the napari-cryoet-data-portal and blik plugins.]()
- Convert a TomoTwin embedding DataFrame to Zarr FormatConverts a given DataFrame to a Zarr file format.
- Download a MRC from the CZ CryoET Data PortalDownload a MRC from the CZ CryoET Data Portal.
- Crop a MRC from the CZ CryoET Data PortalCrop a MRC from the CZ CryoET Data Portal.
- Compute basic fixed features for CryoET DataComputes a feature group for cryoET data using skimage.feature.multiscale_basic_features and saves to a Zarr file.
- Process Cropped Data with skimage FeaturesProcesses a crop of the input MRC data and embeddings, computes skimage features, and writes to Zarr.
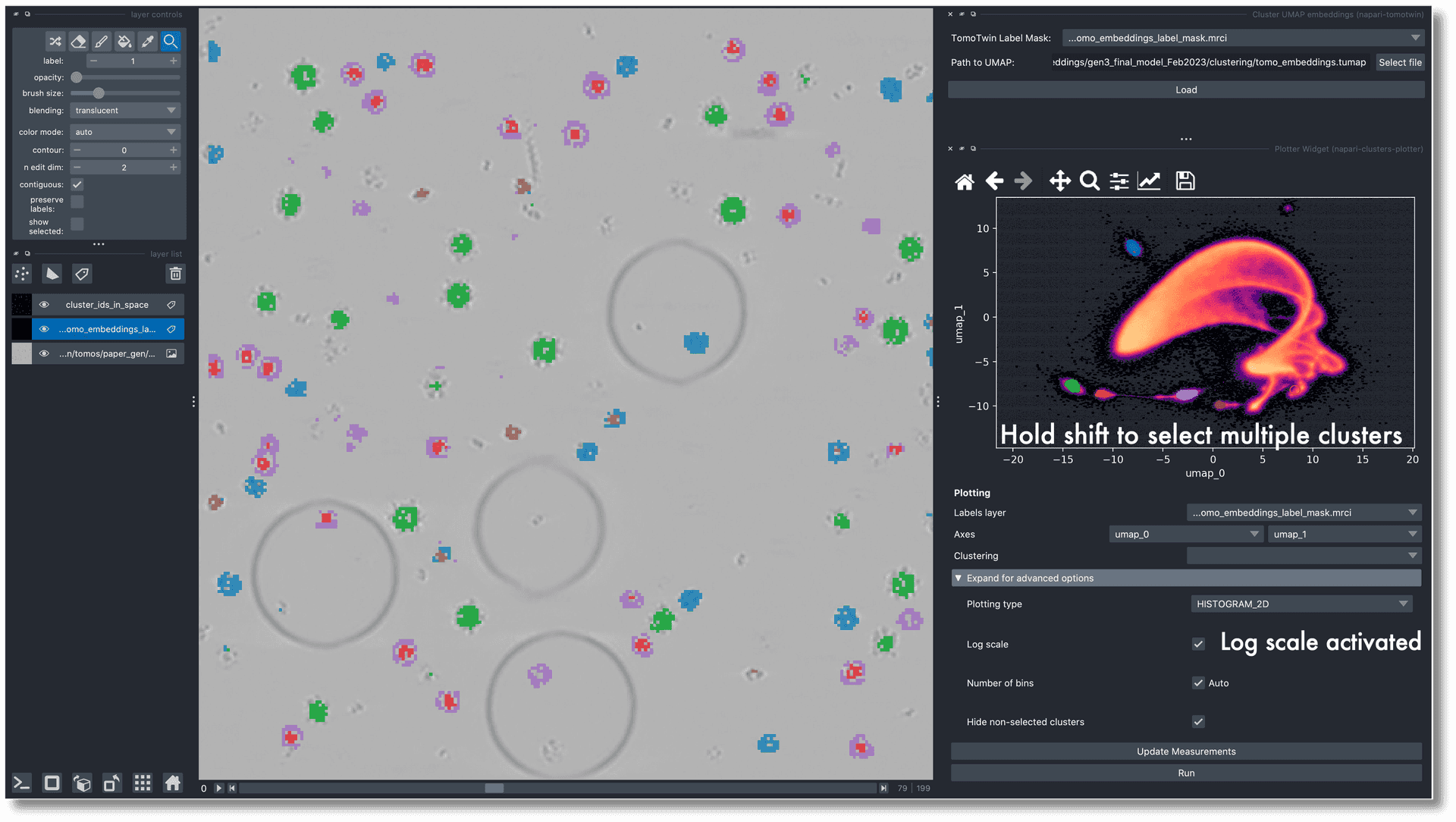
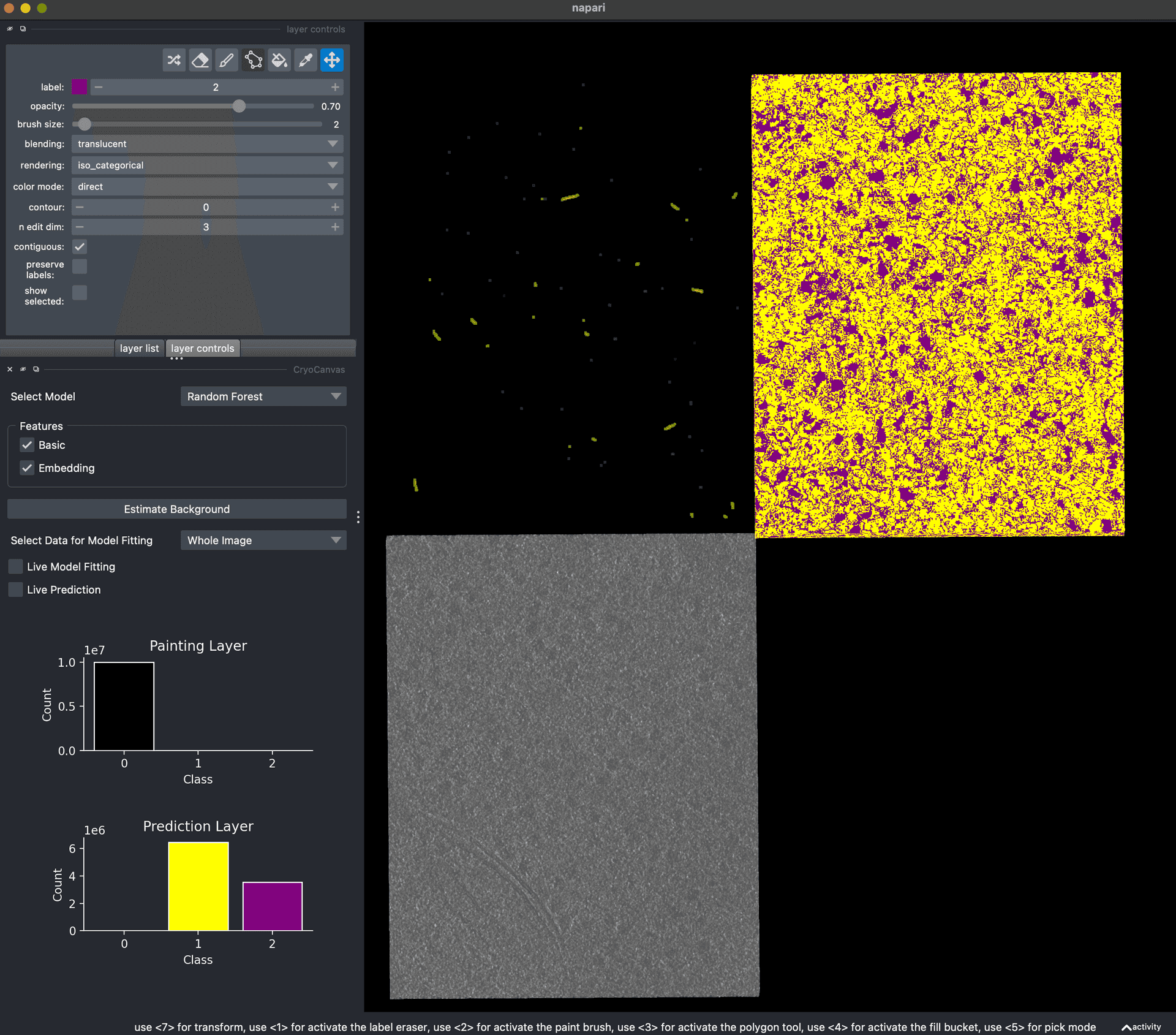
- First portable CryoCanvas demo.First portable CryoCanvas demoKyle Harrington.
![CryoCanvas screenshot.]()
- Generate an embedding with TomoTwin for a crop of a mrcTomoTwin on an example from the czii cryoet dataportal.Rice, G., Wagner, T., Stabrin, M. et al. TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01878-z.
- Generate an embedding with TomoTwin for a Zarr fileTomoTwin on an example from the czii cryoet dataportal.Rice, G., Wagner, T., Stabrin, M. et al. TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods (2023). https://doi.org/10.1038/s41592-023-01878-z.
- Generate Embeddings with Student ModelUse a distilled student model to generate embeddings for a Zarr dataset.Kyle Harrington for CellCanvas
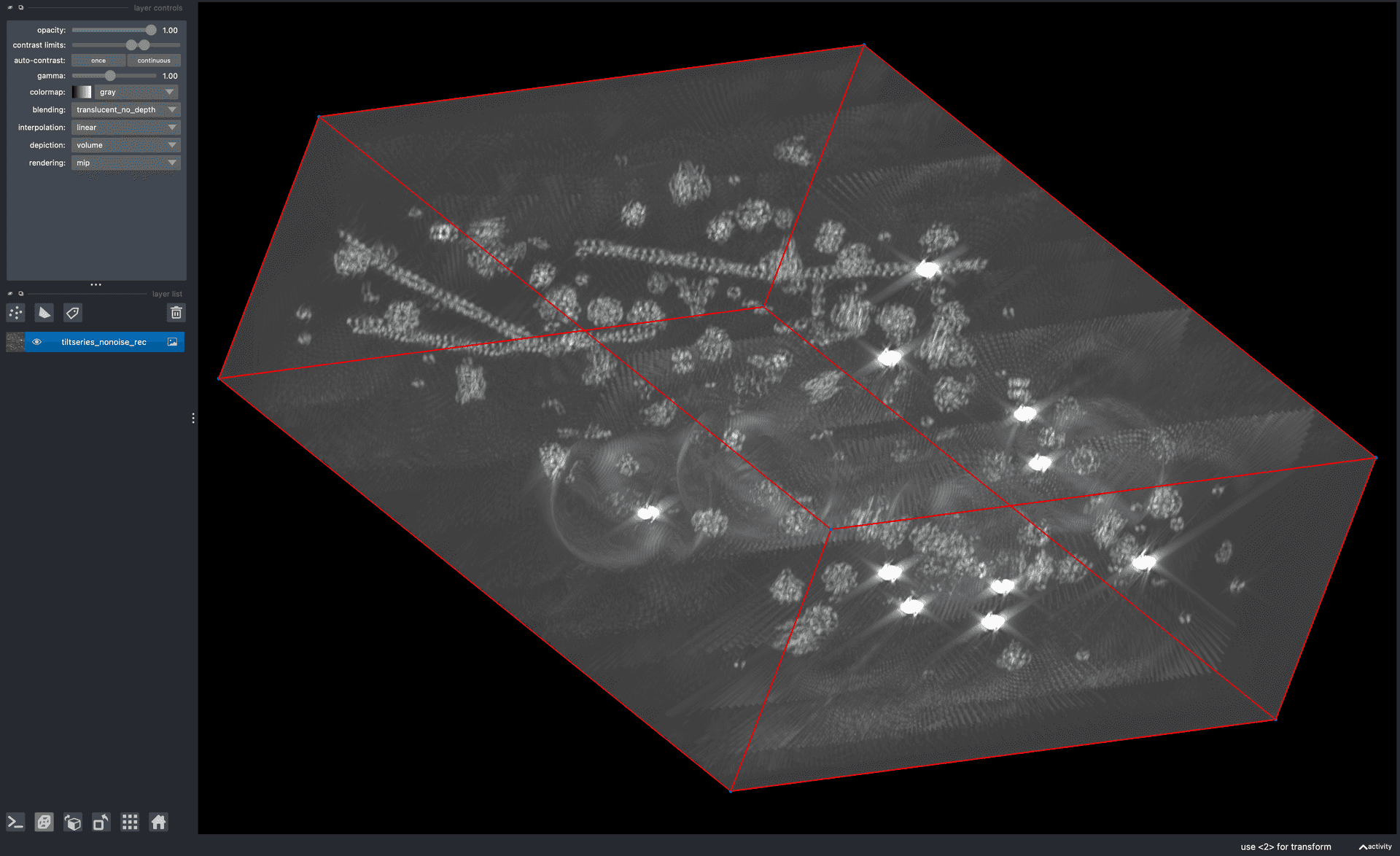
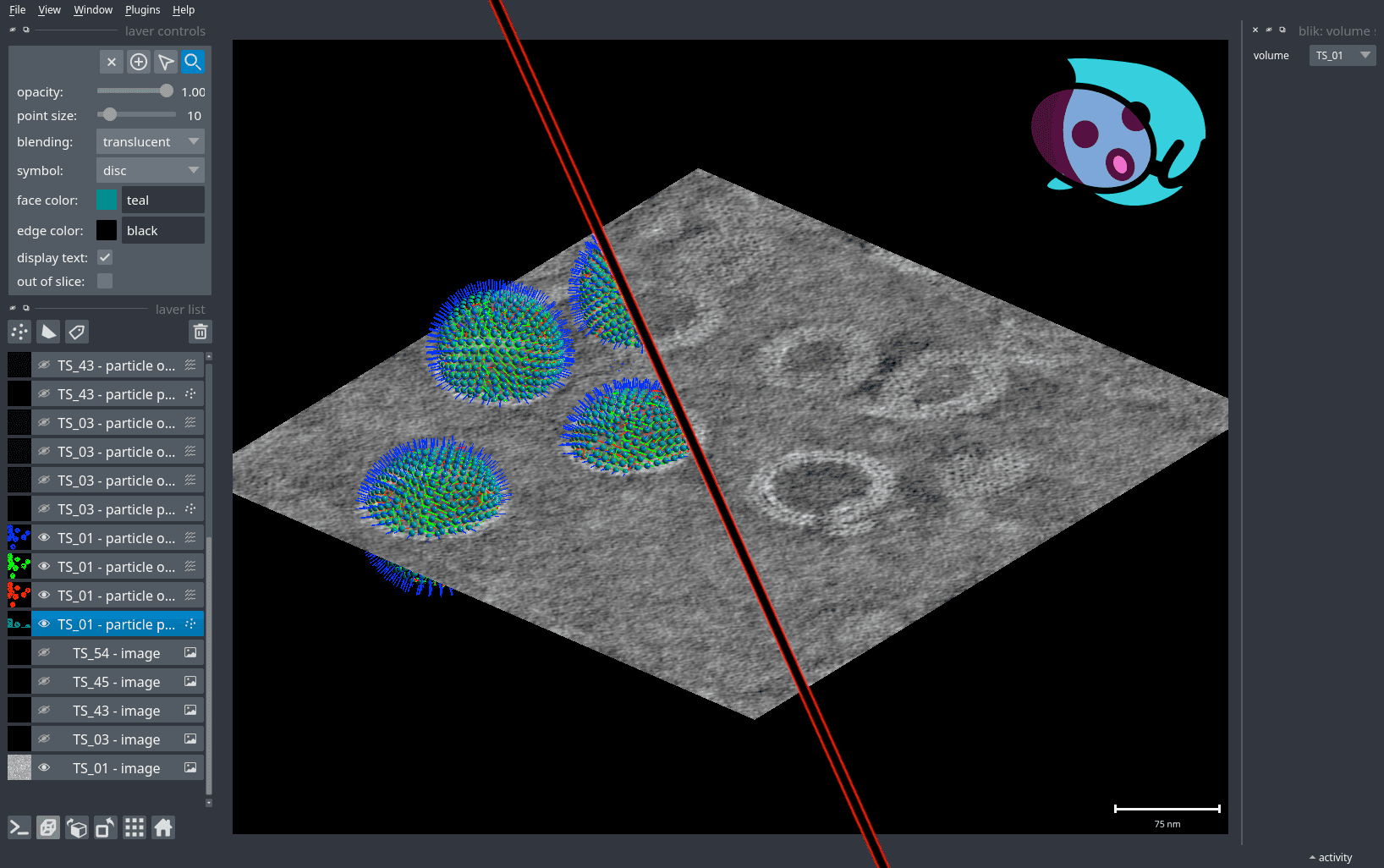
- blik: a Python tool for visualising and interacting with cryo-ET and subtomogram averaging data.blik is a Python tool for visualising and interacting with cryo-ET and subtomogram averaging data.Gaifas, L., Timmins, J. and Gutsche, I., 2023. blik: an extensible napari plugin for cryo-ET data visualisation, annotation and analysis. bioRxiv, pp.2023-12..
![Screenshot of blik.]()
- Compute TomoTwin Embedding and Store with Cropped MRC in ZarrComputes TomoTwin embeddings for a specified crop of MRC data with edge padding and stores both the cropped data and embeddings in a Zarr file.
- Setup a demo environment for CryoCanvasSetup a demo environment for CryoCanvas using the CZ Cryo ET Data Portal.
- PDB Downloader for CATHDBDownloads CATHDB and corresponding PDB files into the specified directory.